The influence of the microbiome (the bacteria in your gut) on cavernoma development
N.b. In the US, cavernoma are called cavernous angioma
Introductory text for the non-scientist
Your microbiome is the collection of the billions of bacteria in your gut. There are many different species of bacteria there, and everyone’s microbiome is different.
A cavernoma is an abnormality in the cells that form the blood capillaries in your brain. These cells separate the blood from the brain cells. We know that cavernoma formation is linked to three genes, called CCM1, CCM2 and CCM3. An abnormality (mutation) in any one of these genes in the cells lining the blood capillaries in the brain is required for the formation of a cavernoma, though a cavernoma does not necessarily form if these genes are abnormal.
Our current understanding of the relationship of the microbiome to cavernoma formation is well described by the three papers in this section.
1. Tang AT et al. (2017) Endothelial TLR4 and the microbiome drive cerebral cavernous malformations. Nature 545 305
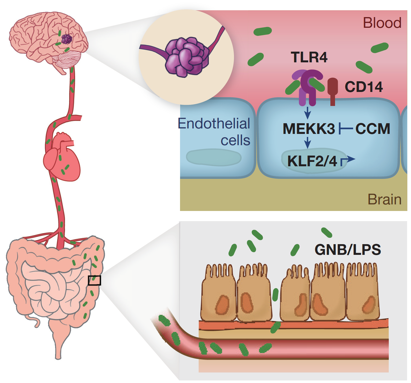 The research reported in this paper is undertaken in mice in which either of the genes CCM1 or CCM2 is deleted in the cells forming the blood capillaries.
The research reported in this paper is undertaken in mice in which either of the genes CCM1 or CCM2 is deleted in the cells forming the blood capillaries.
The paper shows that cavernoma formation in these mice depends on the nature of the gut microbiome. Mice with some microbiomes can form cavernoma and those with other microbiomes cannot do so. A critical difference was the role of a class of bacteria termed gram-negative bacteria (GNB). A potential link to cavernoma in the brain is the production by GNB of a molecule, a lipopolysaccharide (LPS). Evidence from elsewhere has shown that LPS acts on a receptor in cell walls known as TLR4, and in this paper the researchers showed that disruption of TLR4 resulted in cavernoma formation in these CCM1 or CCM2 deficient mice.
The paper shows that deficient genes are not sufficient for a cavernoma to form and that the nature of the microbiome is critical. The paper presents a plausible hypothesis for how this happens, via a molecule (LPS) produced by certain gut bacteria (GNB) that can be carried in the blood to the blood capillary cell walls in the brain, and then trigger a receptor (TLR4) to cause cavernoma formation in mice without one of two cavernoma genes.
2. Tang AT et al., (2019) Distinct cellular roles for PDCD10 define a gut-brain axis in cerebral cavernous malformation Sci. Transl. Med. 11, eaaw3521 (2019)
The research reported in this paper is partly undertaken in mice in which the CCM3 gene is deleted in the cells lining the brain blood capillaries, and partly in humans. In humans, people with the familial form of CCM3 (also known as PDCD10) develop cavernoma decades earlier than those with CCM1 or CCM2 mutations and are more likely to suffer disabling brain hemorrhage and stroke.
The paper showed that the effect of the absence of CCM3 in the blood capillary cells in mice was similar to that caused by the absence of CCM1 and CCM2, implying that the cause of more aggressive action of CCM3 mutations in humans must be elsewhere.
Comparing the microbiome in humans by analysing faecal samples showed a significant difference between the microbiomes of those with familial CCM3 (fCCM3) and those without. However, the microbiomes of those with fCCM3 were indistinguishable from those with fCM1 and fCCM2 and thus the more aggressive nature of CCM3 is not due to the microbiome itself.
What the paper did show is that abnormal CCM3 in mice reduces mucus formation. Mucus forms a barrier in the lining of the gut that helps prevent gut bacteria crossing from the gut to the blood stream. Further, they showed that other mechanisms that reduce the mucus lining also caused an increase in cavernoma formation. One such mechanism is the action of emulsifiers, and the authors showed that the use of one emulsifier did indeed result in an increase in cavernoma formation.
You can download the paper here.
3. Polster SP et al. (2020) Permissive microbiome characterizes human subjects with a neurovascular disease cavernous angioma Nature Communications
The work in this paper is on humans. It is an analysis of the bacterial composition of faecal (stool) samples (i.e. of the gut microbiome) of a large number of people, some with cavernoma and some from the general population. The paper showed that the gut microbiomes from people with cavernoma were distinct from those without cavernoma with the relative abundance of three common bacterial species distinguishing the two with high sensitivity and specificity. Further, the microbiomes of those with sporadic and familial cavernoma and those with single or multiple cavernoma were indistinguishable.
The differences in the microbiomes were consistent with an increase in bacteria enhancing gut inflammation in cavernoma patients.
The paper also showed that cavernoma patients had a higher synthesis of lipopolysaccharide molecules, consistent with the studies on mice described above.
You can download the paper here.
The overall conclusion from the three papers above is that formation of cavernoma in humans is associated with a distinctive bacterial composition in the gut (the gut microbiome) which are inflammatory on the lining of the gut and which increase the synthesis of lipopolysaccharides. Studies in mice have shown that a requisite for cavernoma formation is a gut microbiome which has these same properties. In mice, and hypothesised in humans, either the bacteria themselves or the LPS passes into the blood stream and activates receptors on the blood capillary epithelial cell walls in the brain triggering the formation of cavernoma in mice in which those cells also have aberrant forms of any one of the cavernoma genes, CCM1, CCM2 or CCM3.
That the microbiome is the cause, and not the consequence, of cavernoma formation is shown by the finding that the microbiome causes the activation of the lipopolysaccharide synthesis genes in humans.
Work in mice has shown that an explanation for the greater severity of CCM3 cavernoma is due to the dual action of CCM3 on the lining of the gut and in triggering cavernoma in the brain.
The authors of the papers emphasise that the potential for probiotic treatment or faecal transplants requires considerable further work since trying to alter levels of one or two types of bacteria could lead to other kinds of health problems.

