Table of Contents
Webinar Video
Introduction
We have known for many years that three genes are linked to the formation of cavernoma. Over 99.9% of the population have the normal form of those genes, responsible for tight junctions (TJ) between blood capillary cells. However, less than 0.1% of the population have a mutant form of one of those genes. As described in Part 2, a cavernoma can only form after both copies in a cell of any one of those genes has the mutant form.
Recent research reported in this blog has shown that an additional factor controls the loss of tight junctions: activation of a particular receptor for lipopolysaccharide (LPS) on the membrane of the capillary cell in the brain, known as a TLR4 receptor.
The source of the LPS is certain species of bacteria in the gut that have this LPS in their cell wall. Normally the lining of the gut prevents bacteria crossing into the adjacent blood stream. However, the gut can become leaky if the mucous layer is reduced or absent. When this hakppens, bacteria can escape into the blood stream and reach the brain. If the bacterial outer membrane contains LPS, this can trigger the TLR4 receptor.
It is important that you both read the text and study the figures as you go along. They tell the story together.
How the gut influences cavernoma formation by being the source of a molecule (a lipopolysacchraide [LPS]) which gets into the bloodstream and activates the brain’s blood capillary cells if they carry mutant cavernoma genes
4A. The Gut
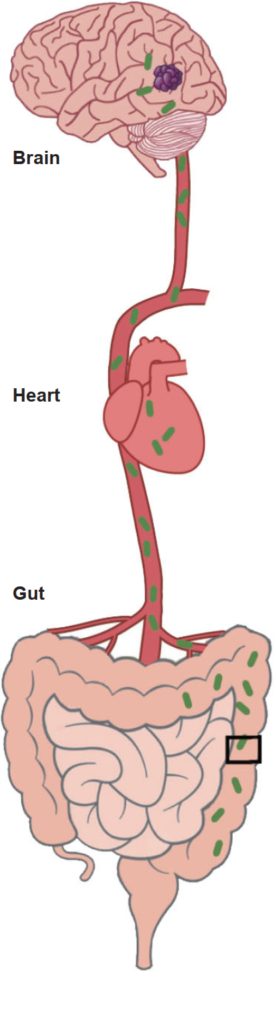
4B. How the gut interacts with the brain: blood circulation
The gut, by which we mean the small and large intestines, has an extensive blood supply receiving nutrients to be distributed around the body. The brain uses a lot of energy and has a very good blood supply too, in order to receive oxygen and nutrients. The heart pumps the blood around the body.
4C. The microbes (mainly bacteriaa) in your gut - the gut microbiome
Your gut is full of bacteria and other microbes, about the same
number (100 trillion) as there are human cells in your body. This microbial content is known as the gut microbiome. There are over 1000 species of bacteria in your gut and everyone has a different mix.
4D. The Gut
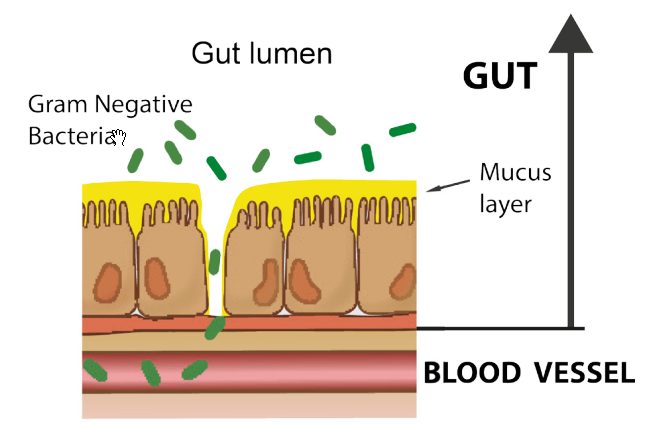
The wall of the gut is lined by several different types of cell. These include goblet cells that secrete the mucus layer that normally covers the cells of the gut wall.
The mucus layer is one barrier to bacteria passing through into the bloodstream, and factors that make the mucus layer thinner make leakiness more likely.
4E. How to explain the unexpected
Much of the basic research on cavernoma is done on what are known as ‘animal models’. These are other species that can develop cavernoma. Mice are commonly used, and there are several breeds of mice that develop cavernoma.

Mark Kahn’s laboratory at the University of
Pennsylvania study the biochemical reactions within the blood capillary cells of mice, and were required to change the building they worked in. The breed of mice they used that developed cavernoma in the old building did not do so in the new one. Same breed, same diet. Same protocols. What had changed?
4F. What had changed?
As Mark Kahn describes: “When we changed the mice’s environment, the bacteria in their guts changed. It wasn’t a deliberate experiment, it was serendipitous. But it had a profound effect. When we sequenced the types of bacteria in their gut, we discovered a change in one type of bacteria. They still had the deleted genes, but they weren’t developing lesions.”
This finding has resulted in a range of experiments on mice first, and then on humans, to elucidate the link between the microbiome and the development of cavernoma. The link is a biochemical, a lipopolysaccharide (LPS)
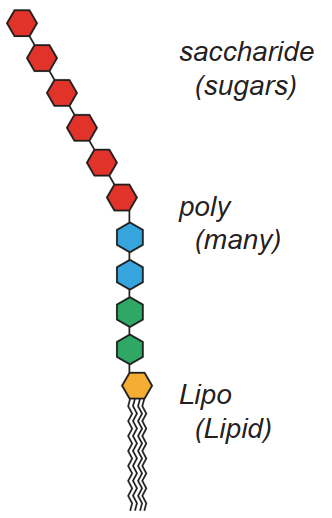
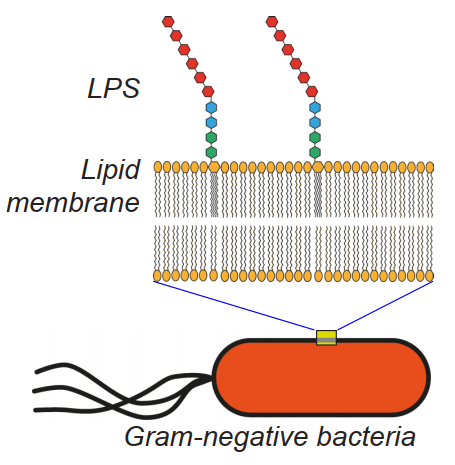
The many species of bacteria in the gut are of one of two classes: gram-negative and gram positive. LPSs are a component of the cell wall of gramnegative bacteria (GNB).
The membrane of bacteria, in common with the membranes of most cells, consists of layers of lipid, and the lipid end of an LPS molecule slots into the membrane of the bacteria as shown.
4G. Membrane Receptor - TLR4 specific for polysaccharides
A membrane receptor is a protein that spans the membrane i.e. it has components either side joined by a part within the membrane. Receptors on the external membrane of a cell have a site that recognises specific molecules in the exterior by binding them to that site. When that site is occupied it causes a change in shape of the receptor on both sides of the membrane.
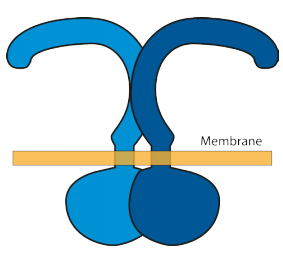
The receptor of interest to this blog has the name TLR4. It is a widespread receptor found in many cell types, and it binds lipopolysaccharide (LPS) molecules. It’s shape changes when it binds LPS, shown schematically in the diagrams. The effect
of binding LPS induces a change of structure in the external part which induces a change internally.
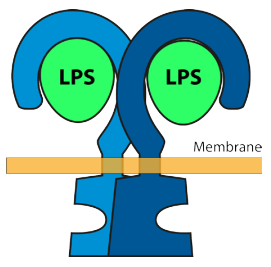
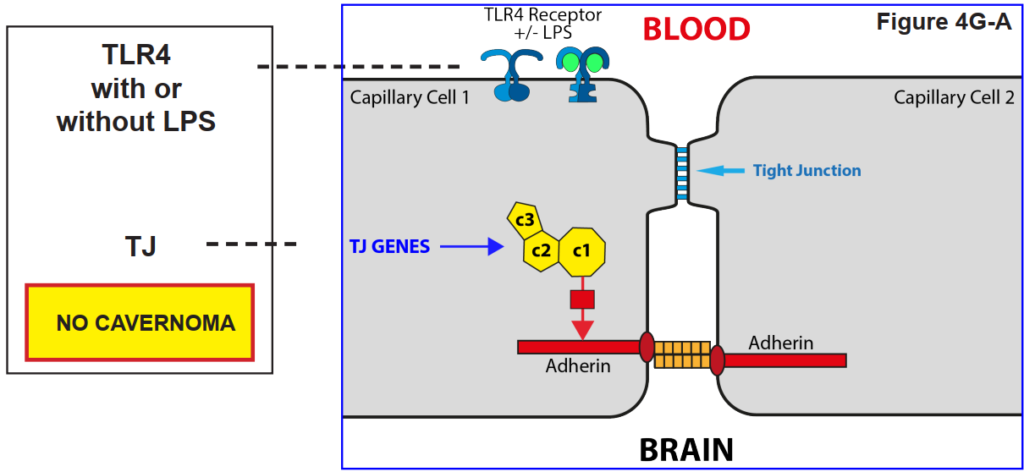
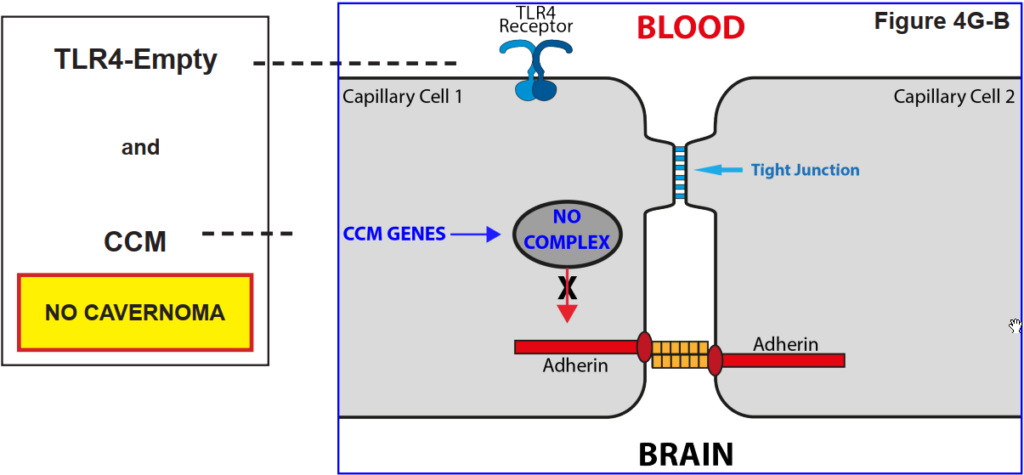
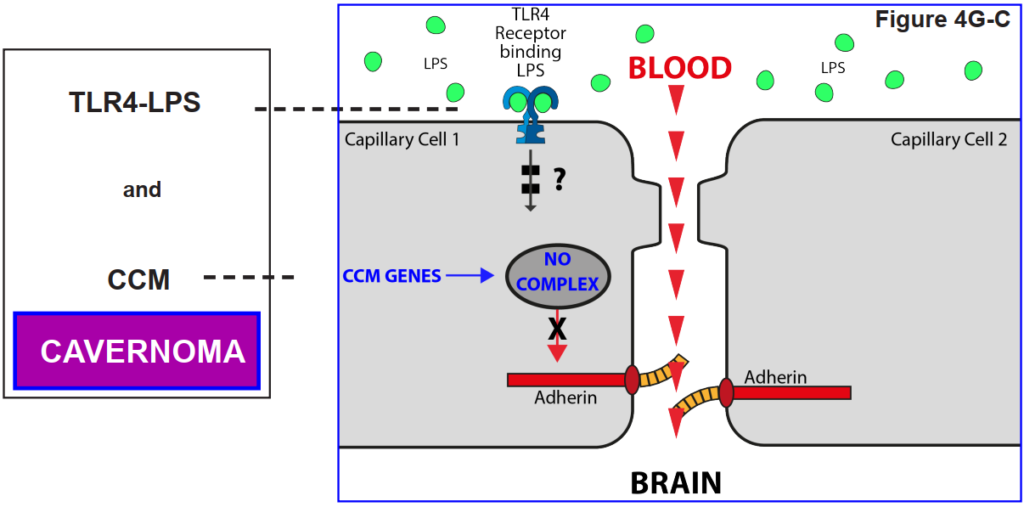
4H. The role of the reception TLR43 on the tight junction
The figures below show that a blood capillary cell does not lose its Tight Junction function unless the TLR4 receptor is activated by LPS.
That is, it was found that a cavernoma can only form provided the capillary cell has both the CCM genes AND the presence of LPS in the
blood stream.
4I. The mix of bacteria in the gut associated with cavernoma
Recent work (2020) analysed bacterial content of the gut from faecal (stool) samples from a large number of people, some with cavernoma and some from the general population. Remember that there are over 1000 species of bacteria in the human gut (the gut microbiome).
Analysis of the bacterial composition showed that:
- A set of the bacteria named the ‘diagnostic set’ shows a clear difference between people with and without cavernoma, and might in future be the basis of a biomarker.
- There is no distinguishable difference between people with different forms of cavernoma in this diagnostic set, i.e. between familial and sporadic cavernoma or between different genes or with different severity.
- The diagnostic set contains reduced levels of bacteria that protect against gut inflammation (associated with leakiness), and increased levels of those that cause gut inflammation.
- A pathogenic set of the bacteria named the ‘permissive set’ is necessary for cavernoma development.
- A distinct set of bacteria is associated with the severity of the cavernoma, including reduction in species that protect against gus inflammation. One of these species is the sole difference found in the microbiome between familial and sporadic cavernoma.
4J. What influences gut leakiness?
The role of CCM3
Leakiness in the gut wall is dependent primarily on the presence of the mucus layer shown in 4D. Mucus is produced by goblet cells in the lining of the gut. A critical component required for mucus production is the gene CCM3. In contradistinction to the role of CCM3 in blood capillary cells described in earlier blogs, where it acts together with CCM1 and CCM2, its role in mucus production is independent of these. Loss of CCM3 in mice results in reduction of mucus and the gut becomes more leaky. Thus the presence of the mutant form of CCM3 has a dual role (a) in enabling cavernoma formation and (b) in increasing the leakiness of the gut wall, thereby explaining why the severity of cavernoma is more severe in people with fCCM3.
Other factors
Emulsifiers will tend to cause a loss of the mucus layer, and diets low in emulsfiers are recommended. Angioma Alliance has produced a CCM-Healthy Cookbook based in large part on reducing emulifiers, a common ingredient in processed foods.
4K. Summary
What the research behind this blog shows is that having the mutant form of any one of the cavernoma genes, CCM1, CCM2 and CCM3, is a necessary but insufficient condition for the loss of the tight junctions between blood capillary cells and cavernoma formation.
Another trigger is also required. This additional requirement is the activation of the TLR4 receptor on the capillary cell membrane by a lipopolysaccharide (LPS) molecule.
The source of LPS is Gram Negative Bacteria (GNB) in the gut, The lining of the gut, including the mucus layer, is normally impervious to bacteria. However, if the mucus layer is compromised the gut wall becomes leaky and bacteria can pass into the blood stream and from there reach the blood capillaries in the brain. The LPS can then activate the TLR4 receptors.
How this then causes the loss of control of the adherin protein that maintains the tight junction (TJ) is unknown at the moment, and the subject of current research.
CCM3, acting independently of CCM1 and CCM2, is required for mucus production. Hence people with the mutant (CCM) form of CCM3 experience more severe forms of cavernoma due to the reduction of the mucus layer. Emulsifiers in food are another factor that can reduce the mucus layer.
Acknowledgements
The figures in 4A and 4B are reproduced with permission from Tang et al (2017), Nature Vol 545 p305. The figure in 4B has been modified to show the mucous layer.
The figure in 4C is reproduced from Lukiw WJ (2020) Front. Cell. Infect. Microbiol., 12 February 2020 | https://doi.org/10.3389/fcimb.2020.00022. This is an open access article distributed under the terms of the Creative Commons Attribution License